DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology
Por um escritor misterioso
Last updated 22 março 2025
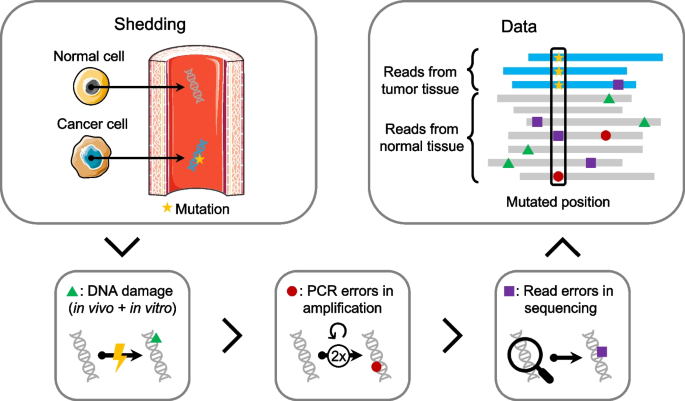
Circulating tumor DNA detection using next-generation sequencing (NGS) data of plasma DNA is promising for cancer identification and characterization. However, the tumor signal in the blood is often low and difficult to distinguish from errors. We present DREAMS (Deep Read-level Modelling of Sequencing-errors) for estimating error rates of individual read positions. Using DREAMS, we develop statistical methods for variant calling (DREAMS-vc) and cancer detection (DREAMS-cc). For evaluation, we generate deep targeted NGS data of matching tumor and plasma DNA from 85 colorectal cancer patients. The DREAMS approach performs better than state-of-the-art methods for variant calling and cancer detection.
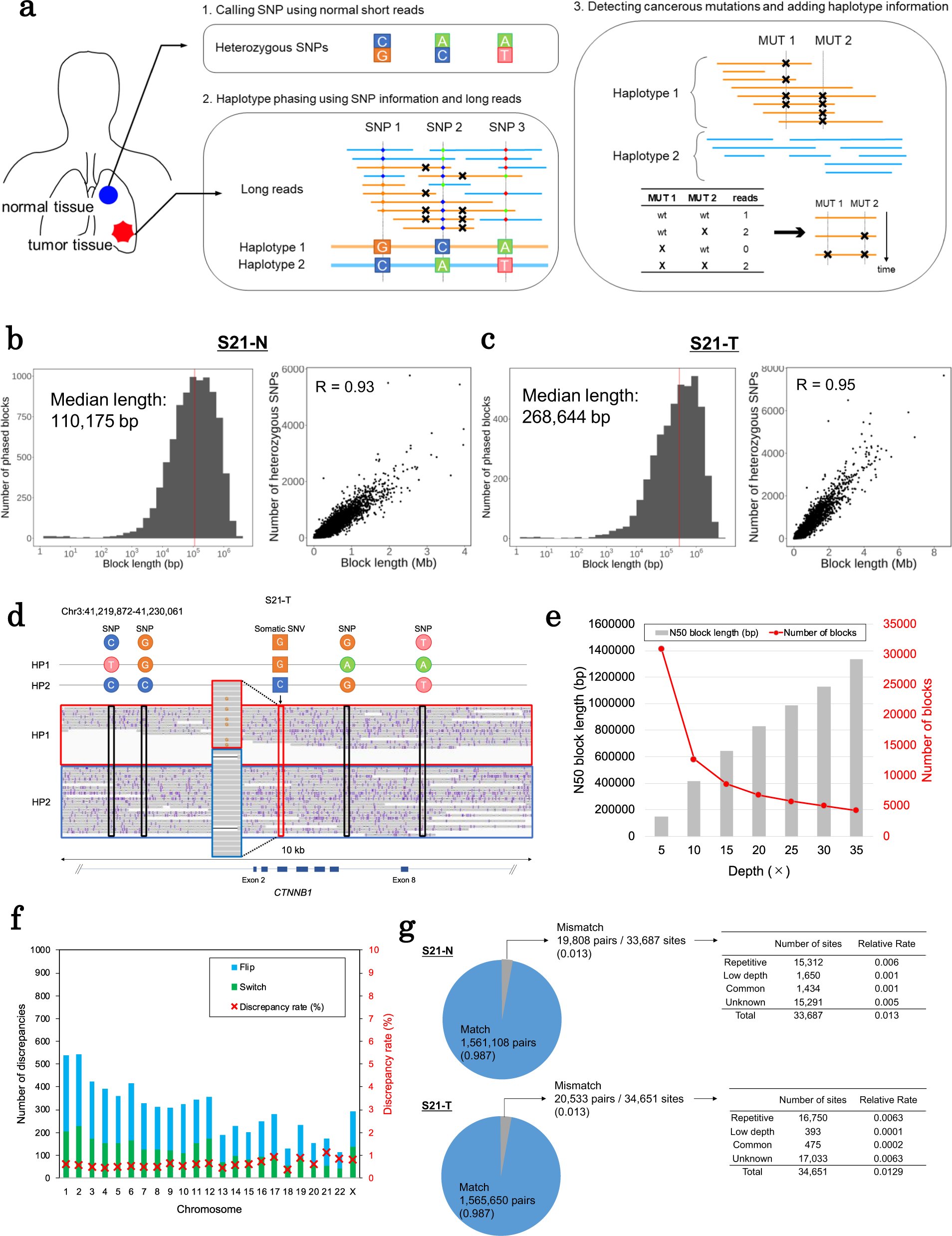
Phasing analysis of lung cancer genomes using a long read

DREAMS: Deep Read-level Error Model for Sequencing data applied to

Applications and analysis of targeted genomic sequencing in cancer
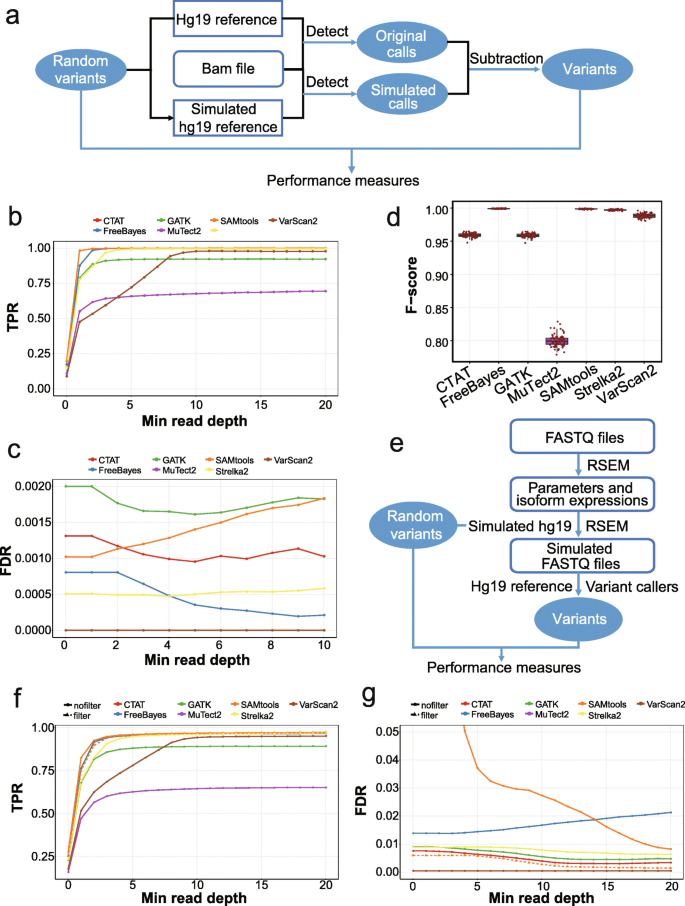
Systematic comparative analysis of single-nucleotide variant

LFMD: detecting low-frequency mutations in high-depth genome
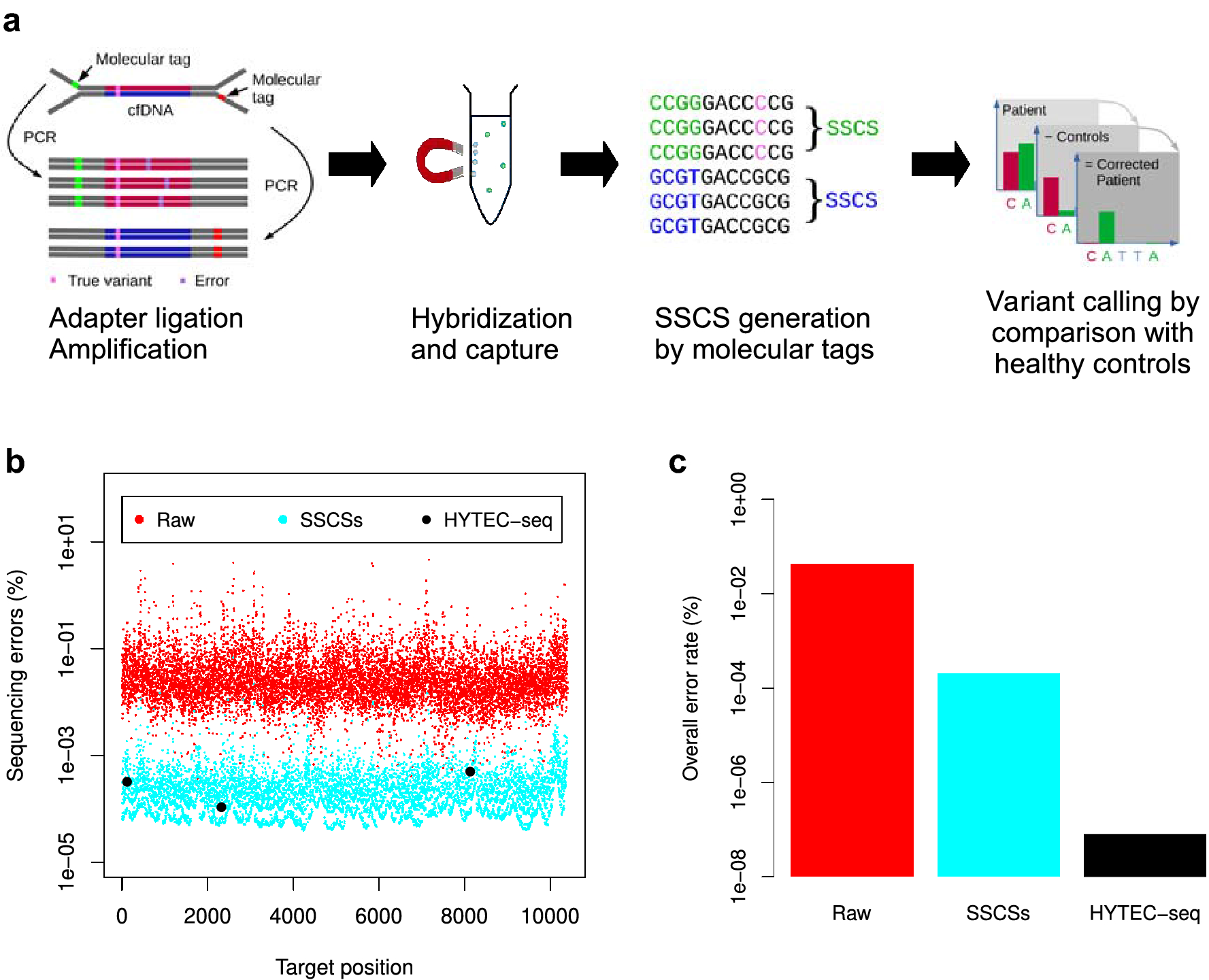
Novel hybridization- and tag-based error-corrected method for

DREAMS: Deep Read-level Error Model for Sequencing data applied to

The changing face of circulating tumor DNA (ctDNA) profiling

DREAMS: Deep Read-level Error Model for Sequencing data applied to

Whole genome error-corrected sequencing for sensitive circulating

DREAMS: Deep Read-level Error Model for Sequencing data applied to

Applications and analysis of targeted genomic sequencing in cancer

Potential error sources in next-generation sequencing workflow. a
Recomendado para você
-
 SCP-7143-J : r/DankMemesFromSite1922 março 2025
SCP-7143-J : r/DankMemesFromSite1922 março 2025 -
![SCP 7143 J THE KNOB [SFW?]](https://i.ytimg.com/vi/qL8GcFVnURE/hq720_2.jpg?sqp=-oaymwEYCI4CEOADSFryq4qpAwoIARUAAIhC0AEB&rs=AOn4CLCHvLM5-ZlLxt7xHLsYYfAnNKRhFA) SCP 7143 J THE KNOB [SFW?]22 março 2025
SCP 7143 J THE KNOB [SFW?]22 março 2025 -
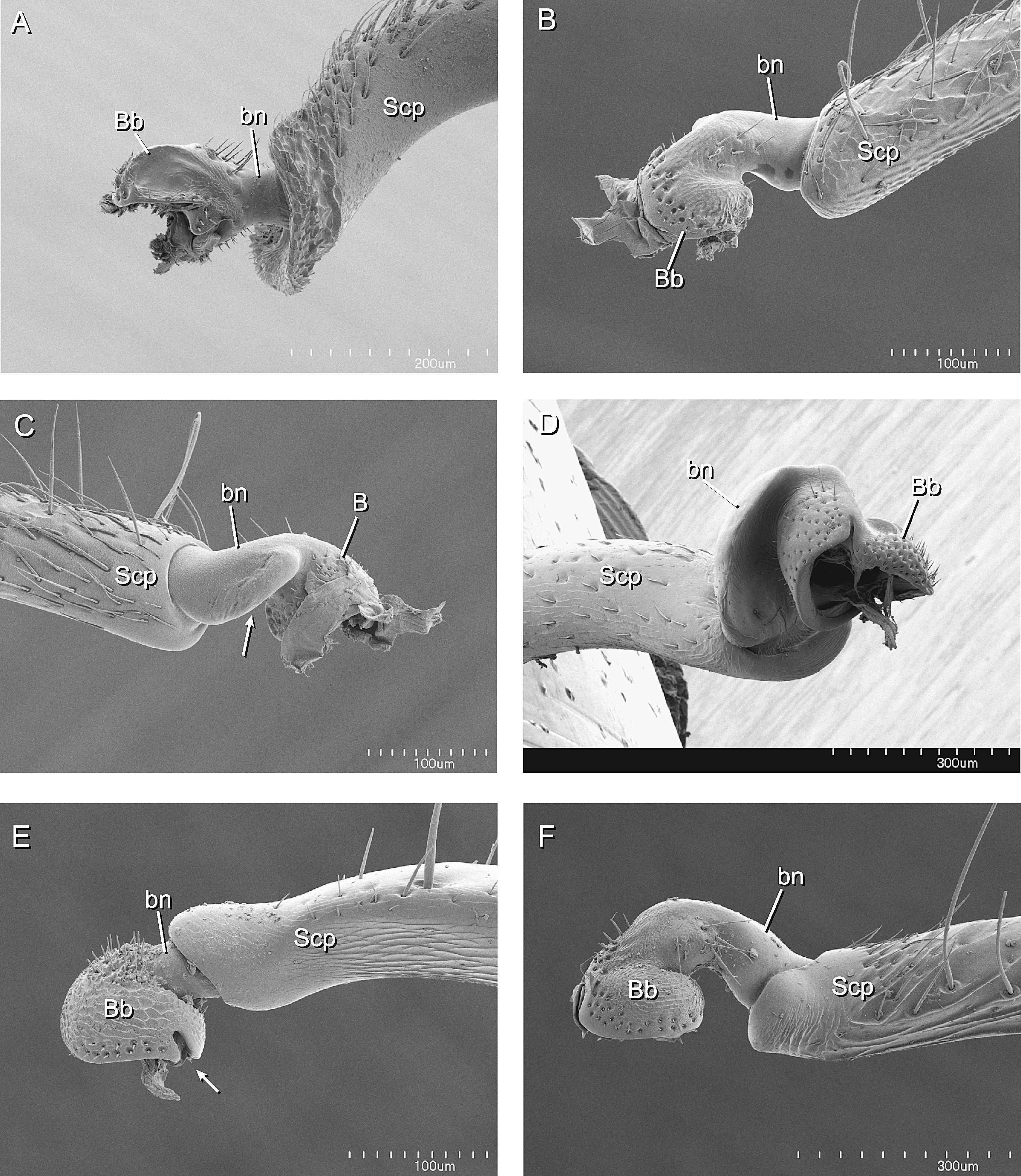 A Phylogenetic Analysis of Ant Morphology (Hymenoptera: Formicidae) with Special Reference to the Poneromorph Subfamilies22 março 2025
A Phylogenetic Analysis of Ant Morphology (Hymenoptera: Formicidae) with Special Reference to the Poneromorph Subfamilies22 março 2025 -
 Pride month logo, SCP Foundation22 março 2025
Pride month logo, SCP Foundation22 março 2025 -
 watched rick and morty thought of this SCP - Imgflip22 março 2025
watched rick and morty thought of this SCP - Imgflip22 março 2025 -
I can't believe this happened 😂 #screammovie #ghostface #scream6 #scr22 março 2025
-
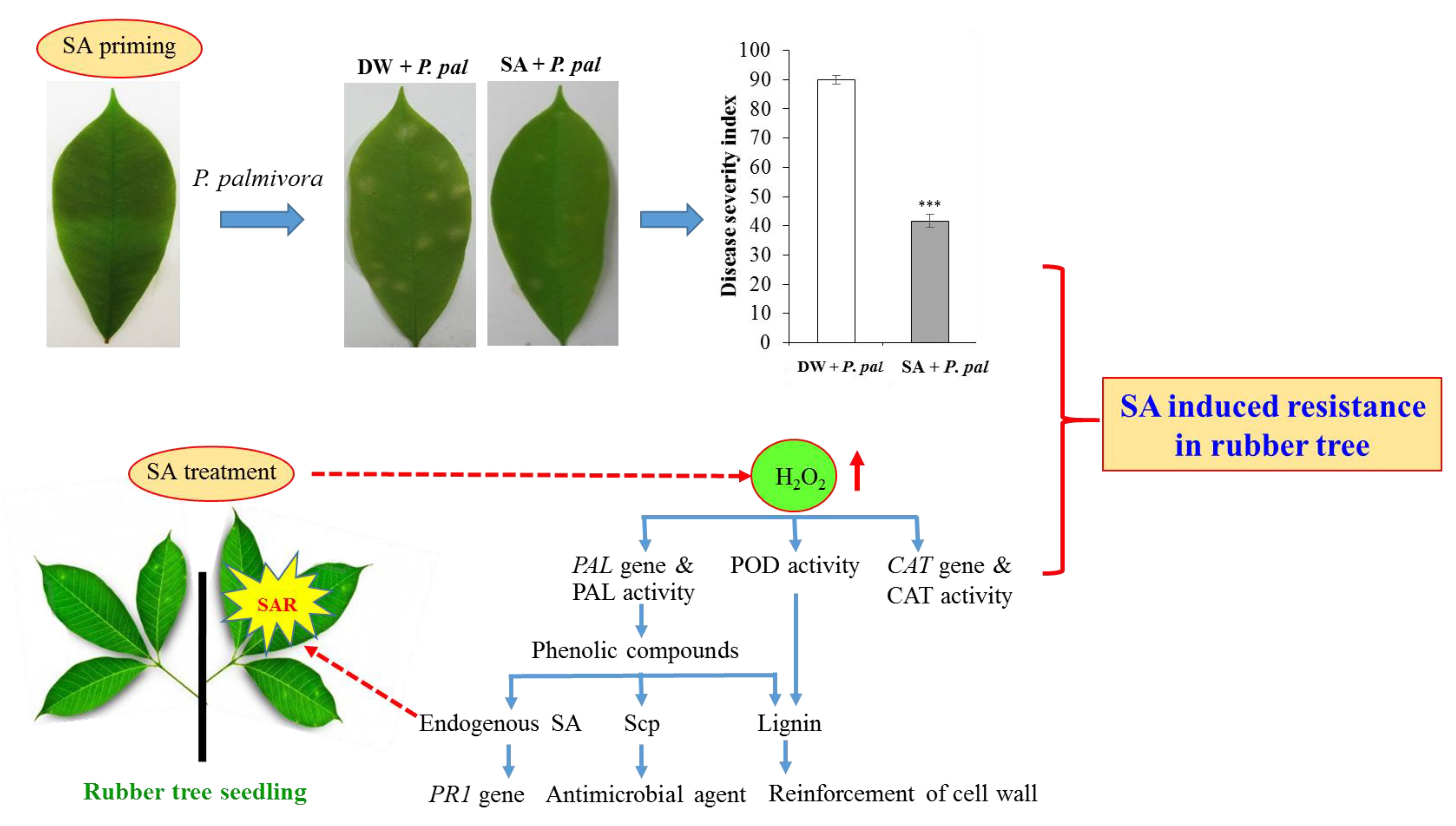 IJMS, Free Full-Text22 março 2025
IJMS, Free Full-Text22 março 2025 -
 Transgenic Mice Expressing Green Fluorescent Protein under the Control of the Melanocortin-4 Receptor Promoter22 março 2025
Transgenic Mice Expressing Green Fluorescent Protein under the Control of the Melanocortin-4 Receptor Promoter22 março 2025 -
 Inhibition of HIV-1 Replication and Activation of RNase L by Phosphorothioate/Phosphodiester 2′,5′-Oligoadenylate Derivatives - ScienceDirect22 março 2025
Inhibition of HIV-1 Replication and Activation of RNase L by Phosphorothioate/Phosphodiester 2′,5′-Oligoadenylate Derivatives - ScienceDirect22 março 2025 -
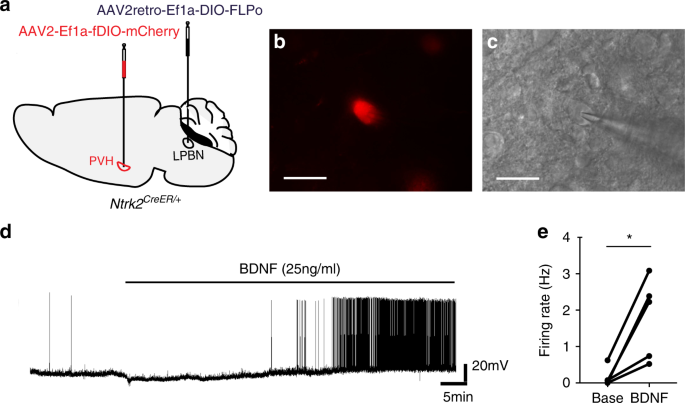 TrkB-expressing paraventricular hypothalamic neurons suppress appetite through multiple neurocircuits22 março 2025
TrkB-expressing paraventricular hypothalamic neurons suppress appetite through multiple neurocircuits22 março 2025
você pode gostar
-
 Metal Sonic cursor – Custom Cursor22 março 2025
Metal Sonic cursor – Custom Cursor22 março 2025 -
 hÉ Tradução de hÉ no Dicionário Infopédia de Francês - Português22 março 2025
hÉ Tradução de hÉ no Dicionário Infopédia de Francês - Português22 março 2025 -
 Sonic Mania Plus Netflix 2024 T-Shirt - Binteez22 março 2025
Sonic Mania Plus Netflix 2024 T-Shirt - Binteez22 março 2025 -
 I Love Emo Boys Fairycore Korean Fashion 2000s Crop Top Girl Yk2 Kawai Gothic Korean Fashion Tshirt - T-shirts - AliExpress22 março 2025
I Love Emo Boys Fairycore Korean Fashion 2000s Crop Top Girl Yk2 Kawai Gothic Korean Fashion Tshirt - T-shirts - AliExpress22 março 2025 -
 Our Rainy Protocol - Anime de eSports recebe trailer e data de22 março 2025
Our Rainy Protocol - Anime de eSports recebe trailer e data de22 março 2025 -
Kiosk Brazil - Requeijão flavored Cheetos available here22 março 2025
-
 One Page Game Websites22 março 2025
One Page Game Websites22 março 2025 -
 MOD: Shotgun king semi weight gain mod v1.1 - Projects - Weight Gaming22 março 2025
MOD: Shotgun king semi weight gain mod v1.1 - Projects - Weight Gaming22 março 2025 -
 Call Of The Night Cotn GIF - Call Of The Night Cotn Yofukashi No22 março 2025
Call Of The Night Cotn GIF - Call Of The Night Cotn Yofukashi No22 março 2025 -
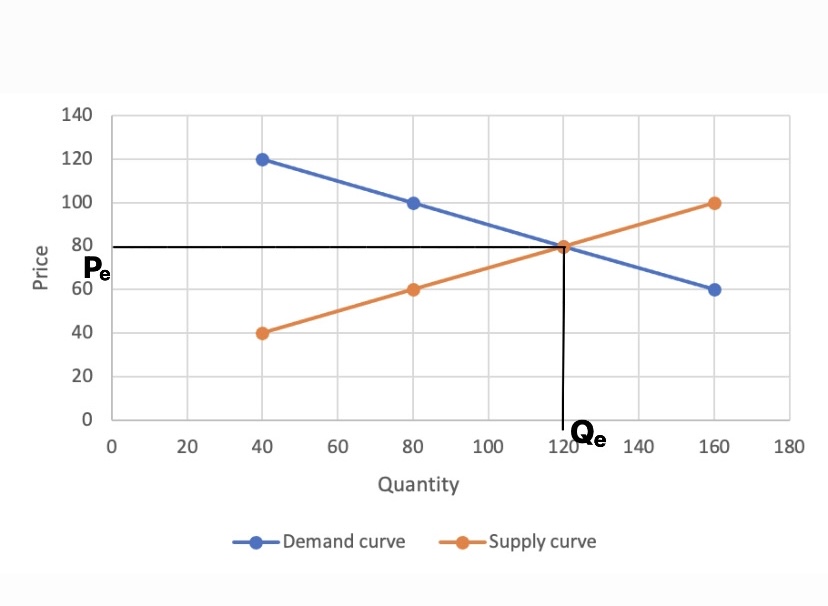 Law of demand - Wikipedia22 março 2025
Law of demand - Wikipedia22 março 2025

